Project List: Published research projects with GitHub repositories:
Genotype-specific responses to iron deficiency in Sorghum
This study highlights the importance of the non-core genome and the utility of the pan-transcriptome in sorghum. Differential expression analysis using the pan-transcriptome revealed 209 genes that responded differently to iron stress in the sweet and non-sweet types, which was more than twice as many genes found using only a single reference genome.
View on GitHub bioRxiv Paper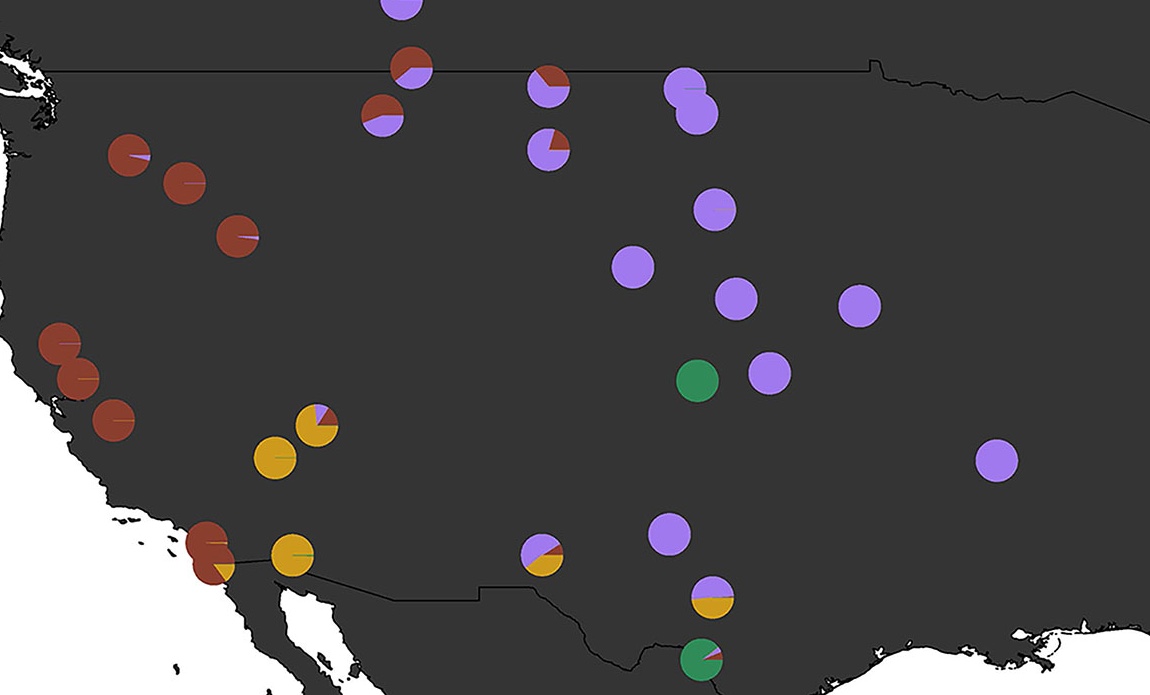
Climate Adaptation and Genetic Differentiation in West Nile virus Mosquitos
Culex tarsalis mosquitoes are a major vector for West Nile Virus and other encephalitis diseases, but despite their growing importance to public health concerns, to date there is a limited number of genetic studies of this species. Here, we uncover distinct patterns of population structure in Cx. tarsalis that appear to have been shaped by both historical geographic isolation and ongoing local adaptation.
View on GitHub GBE Paper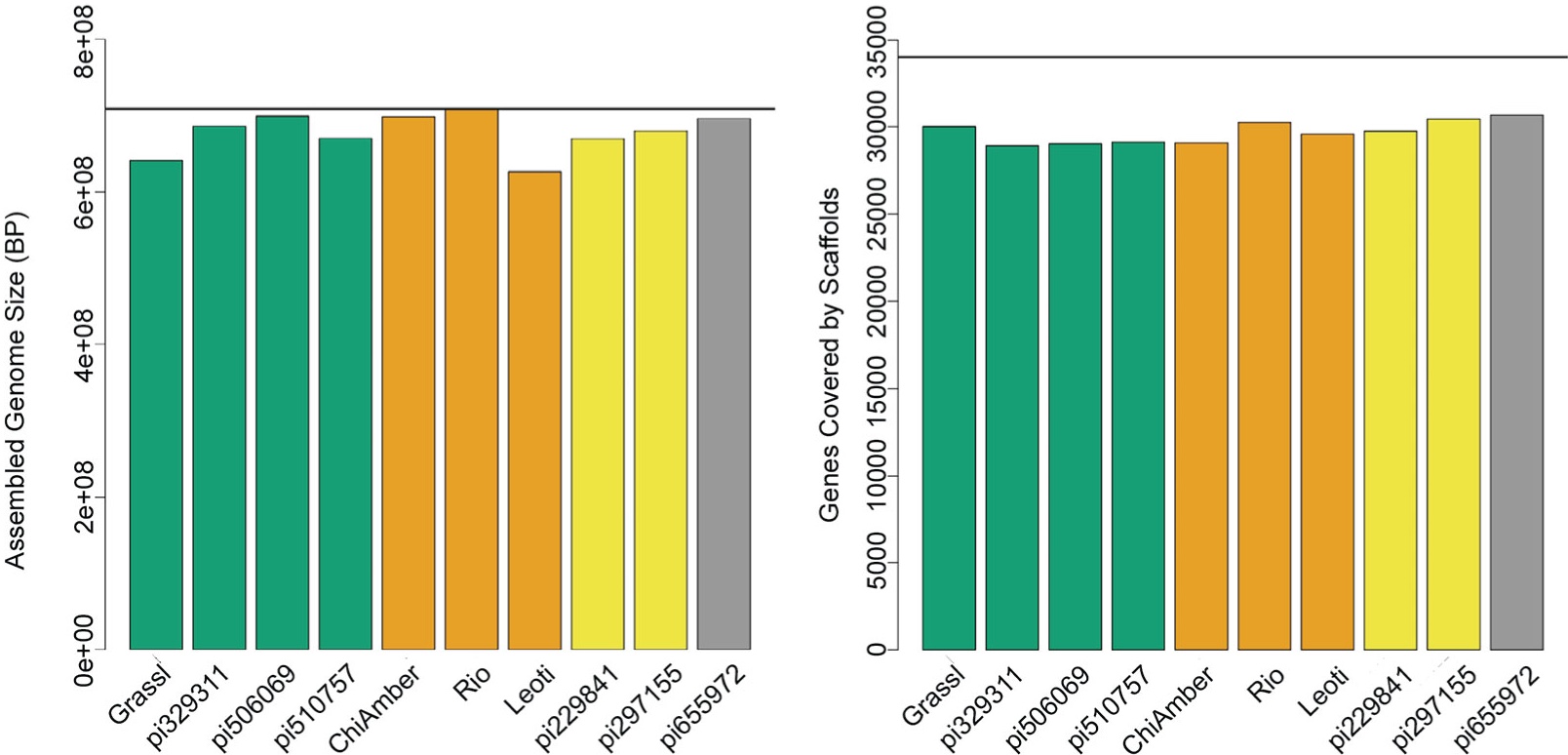
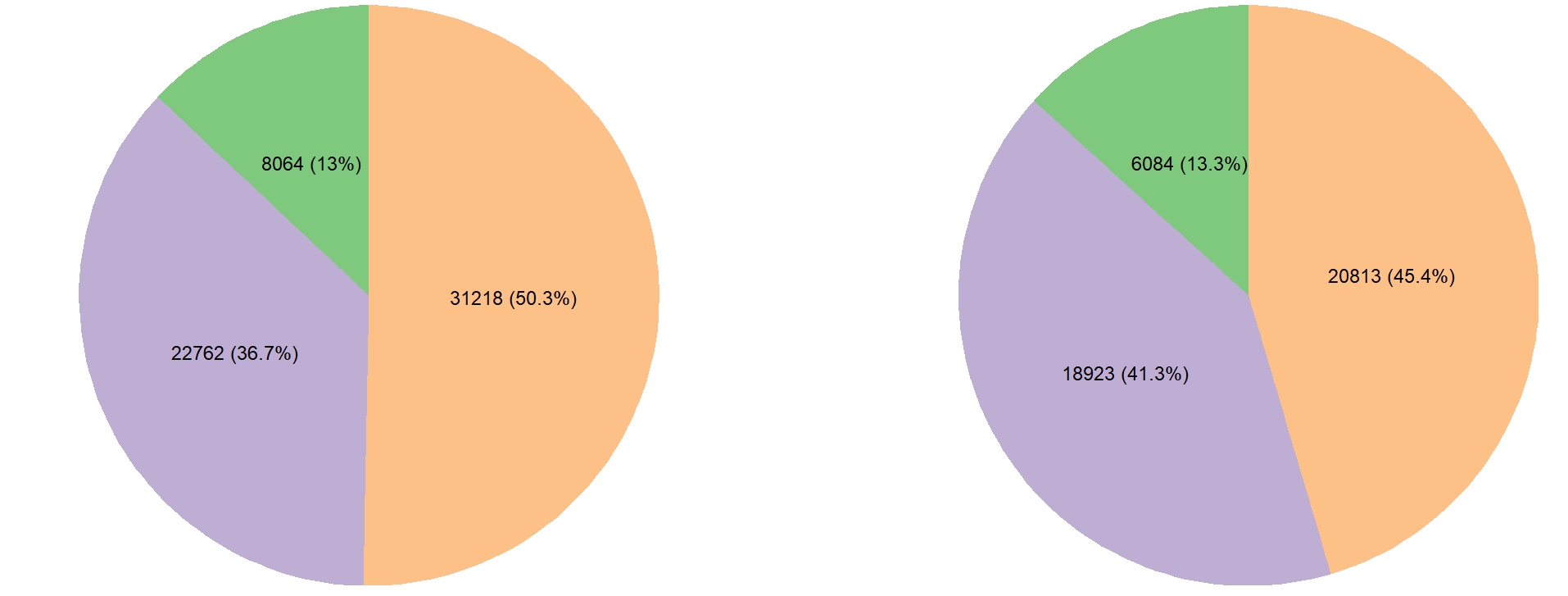
Sorghum Carbon Partitioning Pangenome
Expanding on a recently developed carbon partitioning mapping population, we generated de novo genome assemblies for 10 parental genotypes and identified a comprehensive set of over 24 thousand large structural variants (SVs) and over 10.5 million single nucleotide polymorphisms (SNPs). We show that SVs and nonsynonymous SNPs are enriched in different gene categories and highlight SVs and SNPs occurring in genes and pathways with known associations to critical bioenergy-related phenotypes.
View in SorghumBase Frontiers Paper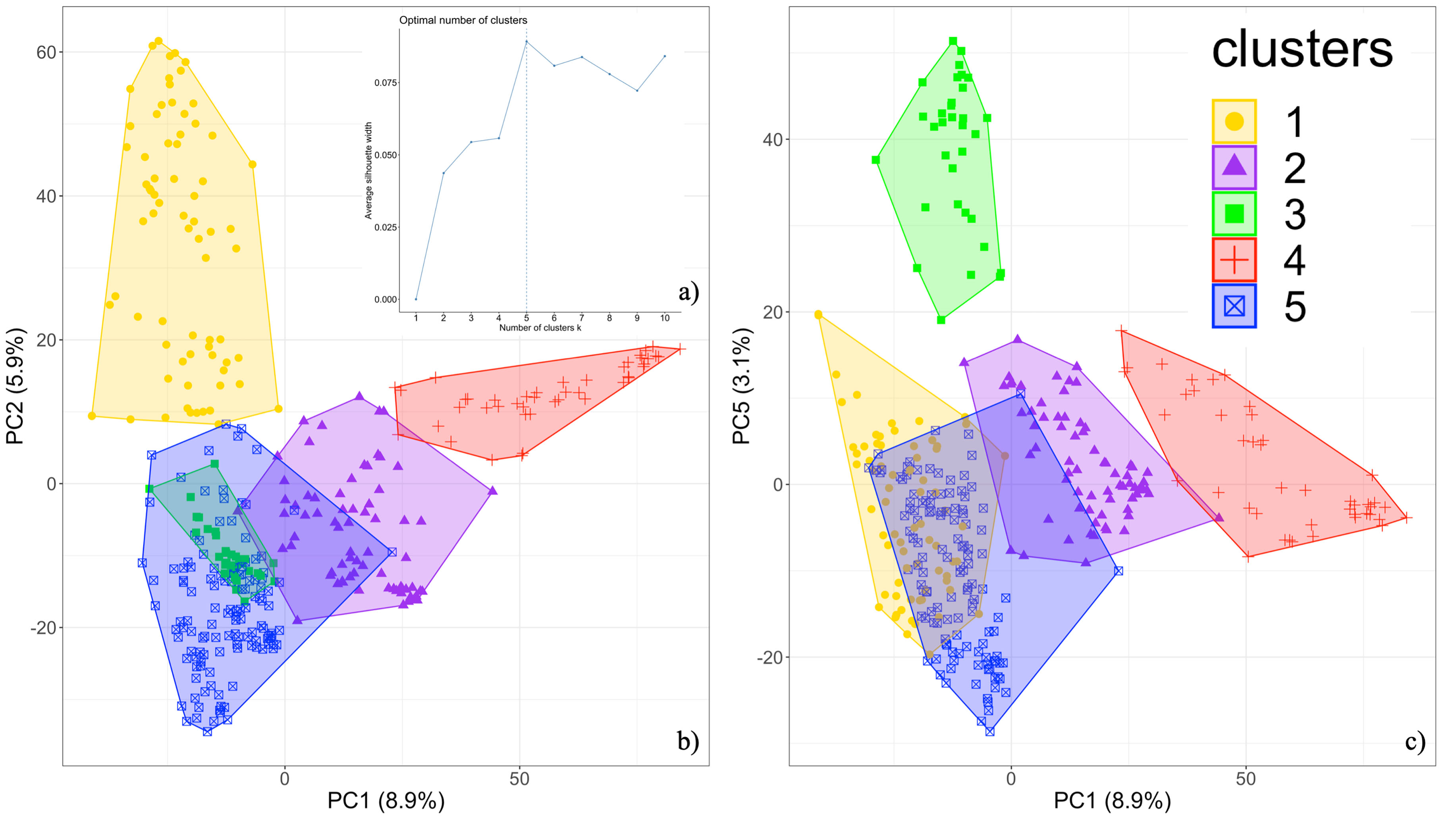
Genomic Patterns of Structural Variation in Sorghum
In this study, we identified genome-wide Structural Variations (SVs) in a collection of 347 diverse sorghum genotypes collected from multiple countries and continents. We show that most SVs appear to be evolving neutrally, but a handful of population-specific SVs are found in genes related to biotic and abiotic stress responses, supporting the possibility that they may contribute to local adaptation.
View on GitHub G3 Paper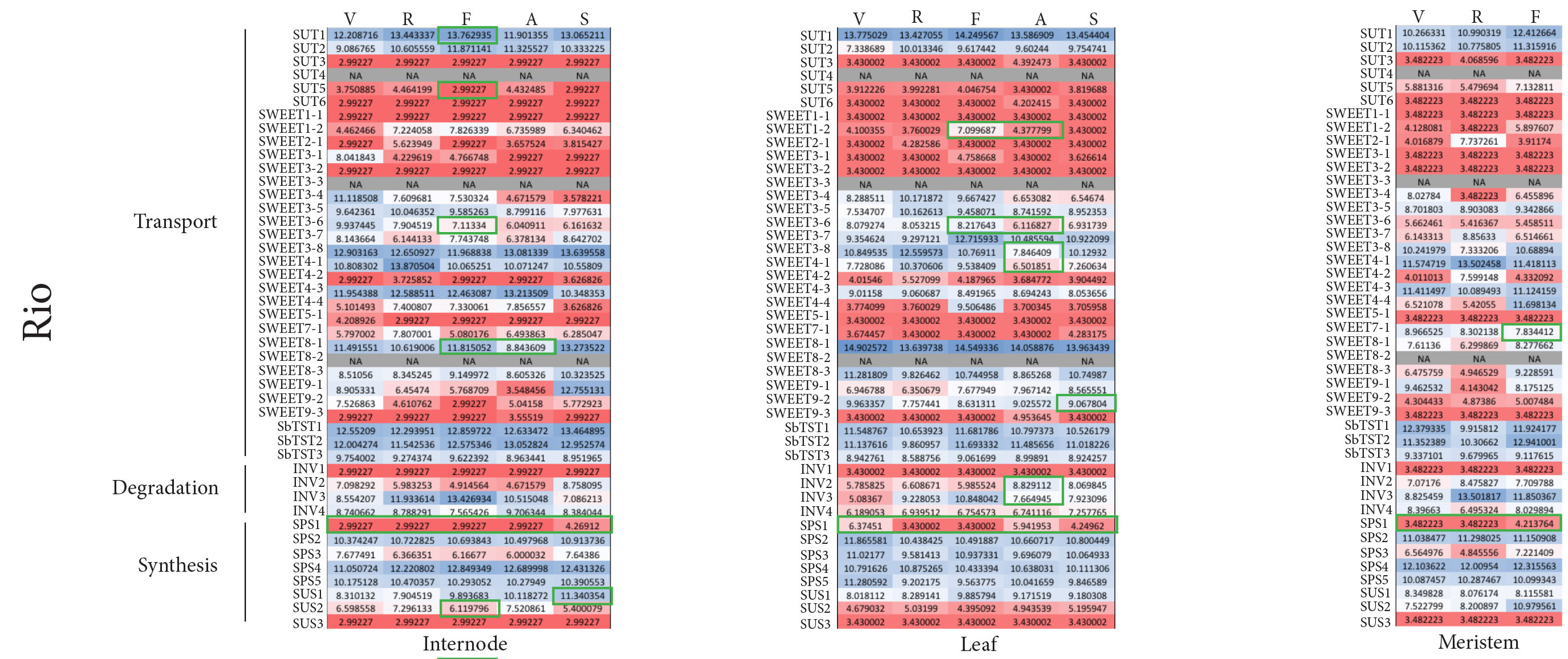
Sweet Sorghum Reference Genome
We generated a new high-quality reference genome for sweet sorghum and compared it to the publicly available grain sorghum genome. We also performed comparative transcriptomics and found that changes in the activity and possibly localization of transporters, along with the timing of sugar metabolism play a critical role in the sweet phenotype.
View on GitHub BMC Genomics Paper
Grain Sorghum QTL Studies
In two separate papers, we use coordinated association and linkage mapping to reveal QTLs that are prime targets to improve both grain quality and grain yield in sorghum. A more robust knowledge of the genetics underlying these complex traits could provide insights into molecular breeding strategies that aim to increase genetic gain.
View on GitHub TAG Paper CropSci Paper
Genetic Divergence in Monarcha Flycatchers of the Solomon Islands
In this study, we used Illumina-sequenced ddRAD tags to identify genomewide patterns of differentiation in three recently diverged island populations of the Monarcha castaneiventris flycatcher of the Solomon Islands. We found that a phylogenetic analysis based on these SNPs produced a taxonomy wherein the two melanic populations appear to have evolved convergently, rather than from a single common ancestor, in contrast to their original classification as a single subspecies.
View on GitHub View Paper
Genomic Variation in Darwin's Finches
We studied genomic variation at the tips of the Darwin's finch radiation; specifically focusing on polymorphism within, and variation among, three sympatric species of the genus Geospiza. Our results suggest that (i) key adaptive traits are associated with a small fraction of the genome, (ii) SNPs linked to the candidate genes are dispersed throughout the genome, and (iii) micro- and macro-evolutionary variation involve some shared and some unique genomic regions.
View on GitHub View Paper
Bioenergy Sorghum Association Panel
To provide a pivotal resource aimed at developing a comparative understanding of key bioenergy traits in grasses, we have established and characterized an association panel of diverse Sorghum bicolor accessions.
View on GitHub View Paper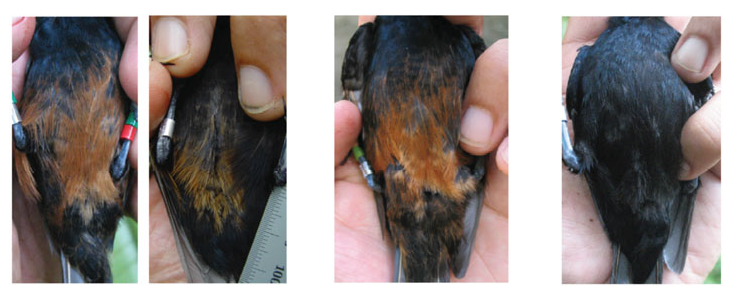
Convergent Plumage Color Alleles
Using a combination of candidate gene and reduced representation genomic sequencing approaches,we explore the genetic basis of and the evolutionary processes that mediate similar plumage colour shared by isolated populations of the Monarcha flycatcher of the Solomon Islands.
View on GitHub View Paper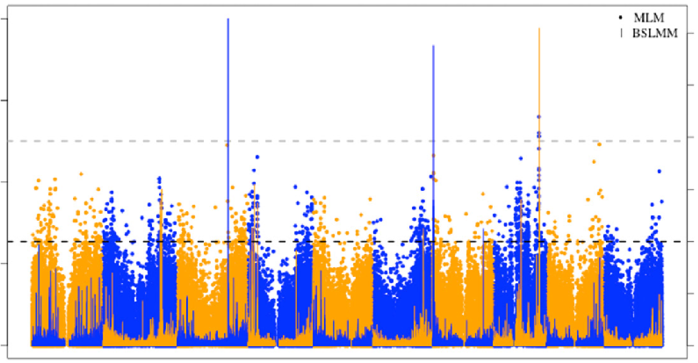
Genome-Wide Association Studies of Grain Yield
Characterization of grain yield per primary panicle (YPP), grain number per primary panicle (GNP), and 1000-grain weight (TGW) in sorghum [ (L.) Moench], a hardy C cereal with a genome size of ∼730 Mb, was implemented in a diversity panel containing 390 accessions.
View on GitHub View Paper